Childbirth pharmacogenetics
Dr.M.Raszek
The miracle of modern day childbirth
Childbirth has become a very orchestrated and standardized process - almost always performed in the safety of a hospital with access to medical professionals who are ready to intervene in any emergencies that might arise. Since more than 99% of all births occur in hospitals, giving birth is the number one reason for hospitalization in Canada which equates to the number one reason for surgical procedures and thus the top reason for total hospital stays by Canadians.
As a consequence of this modernization of the birth process, a dramatic reduction has been observed in maternal mortality and child mortality for nearly the entire globe in the last century: an incredible achievement for humanity and a trend we hope to see continue.
Part of modern medicine’s benefit in the childbirth process is the array of medications available to a mother during labour (depending on her requirements). A client of Merogenomics who has undergone full genome sequencing, later became pregnant and wondered if her collected DNA data could be used in relation to any medications that might be provided to her during the childbirth process. In other words, is there any evidence that person’s DNA could be used to specifically personalize prescriptions and the dosing of drugs used during childbirth? Matching a person’s DNA mutations to how they associate with drug use is a field called pharmacogenetics. Thus to rephrase the question yet another way, is there any pharmacogenetic association data available for childbirth drugs? We decided to do some research.
But first…
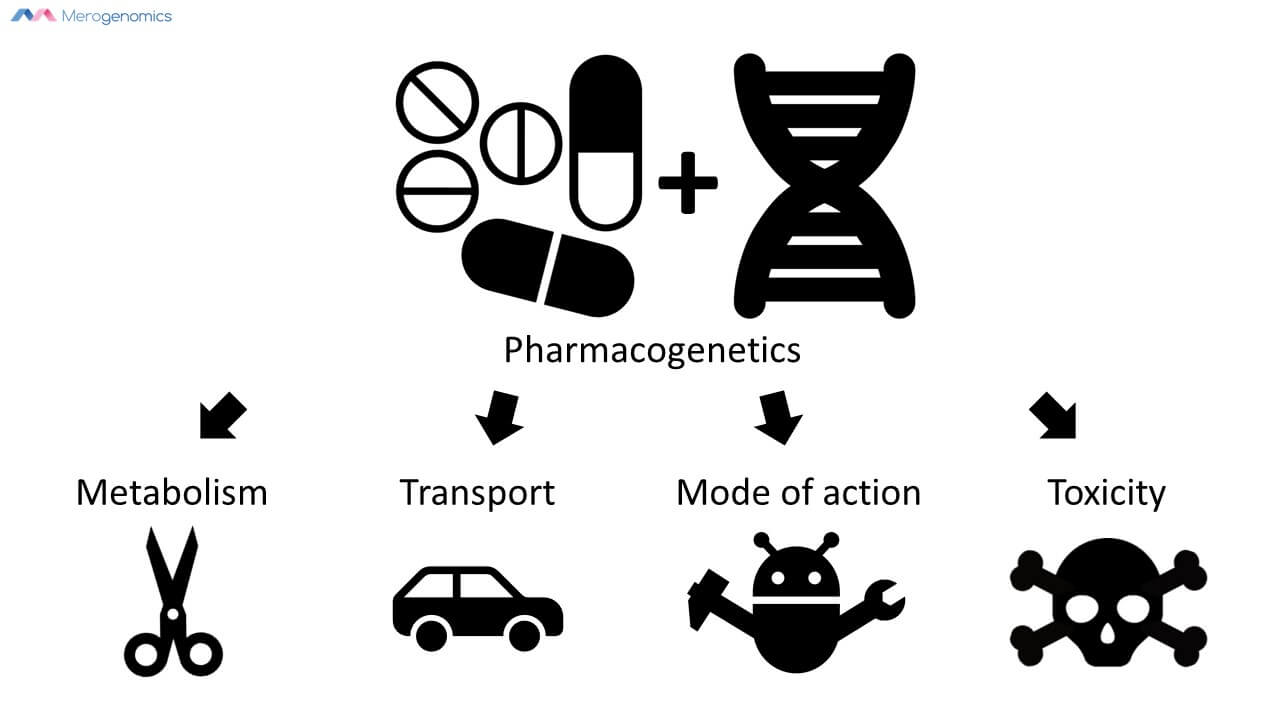
What is pharmacogenetics?
It is a correlation between DNA mutations and how they relate to a person's response to drugs. We are talking about the DNA we inherited from our parents, so there is identical information in all of our cells and thus only needs to be captured once. These DNA mutations can potentially inform us on how drugs can be metabolized and transported; how they might affect the drug’s mode of action; and finally what any toxic response might be. But for the most part, pharmacogenetic information tells us about drug metabolism.
In essence, this allows us to personalize medication use according to someone’s DNA by understanding their built-in biological systems that govern their drug responses. To give you an example, one person can be a fast metabolizer while another person will be slow metabolizer. The fast metabolizer will break down the drug too fast and the drug will have a diminished response. A very slow metabolizer will break the drug down too slow, which could allow the drug to accumulate between doses, potentially exacerbating any negative effects of the drug at that dose. Or, in this case if the medication is a prodrug, meaning the drug has to be broken down first into active metabolites, the reverse will be true and this should also be taken into consideration for correct dosage.
Knowing where someone fits on the scale of drug metabolism allows tailoring of drug dose accordingly. This is probably the most practical use of pharmacogenetics that is observed most frequently in clinical use.
Until now we had no way of knowing this and thus initially, medications have to be given to people somewhat blindly in terms of their expected effect (unless we know some family or personal history of drug reactions). Currently, the only way we would be able to know each drug reaction potential was if we did some complicated blood analysis for every medication. But with DNA sequencing, we can finally get there. And you only need to do a genomic test once in your lifetime.
Of course there are other factors influencing drug utility besides the metabolic enzymes like: diet, other medications being used, infections, and so on. But these complex factors are present at all times. For now, pharmacogenetics is the best we can do. In theory, we will likely even see increased predictability in the future by incorporating these additional complex factors, as we are currently seeing these trends in other areas of DNA sequencing that are being used to predict the impact of such risk factors - but that is quite a complex area of study requiring lots and lots of data (and money).
Does it work? Yes. In fact, pharmacogenetics is an area of DNA sequencing with really well established clinical utility. A database tracking this information, the Clinical Pharmacogenetics Implementation Consortium (CPIC), is one of the best examples of databases that tracks the clinical utility of biomarkers. It is a reputable and extremely well researched medical database. If you are a pharmacist, you will be able to learn lots by accessing it. You can try looking for any medication of interest on it. The evidence is listed from highest to lowest level, and the pharmacogenetic reports are only supposed to report on the two highest levels of accumulated evidence for each medication.
Pharmacogenetics of common drugs used in childbirth
So what were we able to find with regards to childbirth medications? We first obtained the list of the most likely drugs to be used during labour that our client obtained from her obstetrician.
The first table lists these drugs plus what kind of pharmacogenetic information is available on them from the two most authoritative databases that catalog pharmacogenetic data today. These include the Phamacogenomics Knowledge Base (PharmGKB) which assesses the clinical validity of the mutation. Clinical validity refers to the strength of association between the gene and the drug response based on accumulated evidence. The other database is the aforementioned CPIC database which measures the clinical utility of pharmacogenetic information, or in other words, whether the DNA test results actually impact prescribing action (drug avoidance or dosing change).
Of the two, establishing clinical utility evidence is much more difficult because it usually requires complicated experimental designs that are very expensive to carry out. The fact that pharmacogenetic information even has a dedicated database for collecting evidence to establish clinical utility is an amazing achievement and an indicator of how effective personal DNA information can be in driving clinical action. The field of DNA sequencing is undergoing enormous growth right now but many of the proposed medical associations still await some demonstration of having real clinical utility in practice. As mentioned, clinical utility of DNA testing is an ongoing and slow process. The information listed by each of the databases includes how many genes are known to influence the effect of medication as well as how many mutations (otherwise known as variants) within those genes have been described to be affecting the drug outcome plus what level of evidence is available - where the higher the number or the letter of alphabet (depending on which database we are using) the lower the level of supporting evidence that has been accumulated.
In reviewing the databases, most of the pregnancy drugs have no genetic information linked to them at all. Some of the drugs had lots of information listed, but it is usually poor quality with little evidence (sort of like the variants of unknown significance that a person can get when assessing their DNA for health outcomes).
| Purpose | Drug | CPIC listing (utility) | PharmaGKB (validity) | |||||||
| Gene(s) | Evidence level (A-D) | Gene(s) | Variant(s) | Evidence level (1-4) | ||||||
| Induction drugs | Cervidil | |||||||||
| Oxytocin or pitocin | ||||||||||
| Pain management drugs | Epidural anesthesia | |||||||||
| Nitrous oxide | 1 | 2 | 3 | |||||||
| Morphine | 2 | C/D | 13 | 30 | 2B-4 | |||||
| Acetaminophen | 11 | 24 | 3-4 | |||||||
| Hydromorphone (Dilaudid) | ||||||||||
| Fentanyl (Sublimaze) | 2 | C/D | 10 | 25 | 2B-4 | |||||
| Sufentanil citrate (Sufenta) | 3 | 3 | 2B-3 | |||||||
| Nalbuphine hydrochloride (Nubain) | ||||||||||
| Bupivacaine | ||||||||||
| Ropivacaine | ||||||||||
| Epinephrine | ||||||||||
| Naloxone (Narcan) | 1 | C/D | 1 | 1 | 2B | |||||
| Ephedrine | 1 | 1 | 3 | |||||||
| Phenylephrine | ||||||||||
| Symptom management drugs | Dimenhydrinate (Gravol) | |||||||||
| Diphenhydramine (Benadryl) | ||||||||||
| Penicillin | ||||||||||
| Metronidazole (Flagyl) | ||||||||||
The second table dives into the details for some of the drugs that had a level of evidence of 2B in the PharmGKB database. Some of the drugs listed are influenced by the same gene and the same mutation, and that makes sense, as the product of the gene that is involved in drug handling in some way (most often drug metabolism, meaning breaking down the drug into simpler by products) usually will work on multiple chemicals. Remember that a gene is just a DNA blueprint to create some tiny molecular robots inside the cell for some working purpose, for example, robots that chop up chemicals.
| Drug(s) | Gene | CPIC listing evidence level (A-D) | PharmaGKB (validity) | |||||||
| Variant | Evidence level (1-4) | Genotype | Impact | |||||||
| Morphine | - | - | rs2952768 | 2B | C/T | CC may have increased opioid analgesic requirements after surgery as compared to CT or TT | ||||
| Fentanyl | - | 2B | ||||||||
| Morphine | ABCB1 | C/D | rs1045642 | 2B | A/G | AA may have improved response to opioids and may require a decreased dose as compared to AG or GG. Contradicted in some studies | ||||
| Fentanyl | C/D | 2B | ||||||||
| Morphine | COMT | - | rs4680 | 2B | A/G | AA may have an increased response to opioids and may require a lower dose as compared to AG or GG. Contradicted in some studies | ||||
| Sufentanil citrate | - | 2B | ||||||||
| Morphine | OPRM1 | C/D | rs1799971 | 2B | A/G | AA may have improved response to opioids and may require a decreased dose as compared to AG or GG. Contradicted in some studies | ||||
| Fentanyl | C/D | 2B | ||||||||
| Sufentanil citrate | - | 2B | ||||||||
| Naloxone | OPRM1 | C/D | rs1799971 | 2B | A/G | AA may have lower cortisol response to naloxone as compared to patients with the AG or GG | ||||
According to the CPIC database “CPIC level C and D gene/drug pairs are not considered to have adequate evidence or actionability to have prescribing recommendations.”
The PharmGKBdatabase levels of evidence are based on multiple criteria including data replication, statistical significance and study size.
According to these criteria, for a gene to be a pharmacogne, only level 2A evidence is considered and hence more likely to have functional significance. Anything below that threshold remains speculative.
Thus at the moment, none of the medications have enough evidence to know if there would be a specific benefit to a pregnant woman by knowing her genotype for the sites listed in the second table.
However, this is a drop in the sea compared to all the drugs used in pregnancy overall, with the latest count being more than 1200 pregnancy related medications according to most recent reference guide.
The take home message is that childbirth drugs are not yet ready for pharmacogenetic prime time as compared to some other drugs for which validity and perhaps even utility has already been established. If you are considering a good quality pharmacogenetic test then it should be a test that is probing only for DNA information with some established validity or utility or in the least, should provide results only for medications with such established genetic associations. There will be tests out there that will promise you results for hundreds of medications and that should actually be a red flag to you because a pharmacogenetic test that provides results for medications without established validity or utility can be confusing and potentially dangerous.
The above drugs for pregnancy are a good example. As you can see from the information currently available, there is not yet enough evidence for a doctor to be making prescription decisions based on a patient’s DNA data. As a consequence, information related to these medications should not even be listed on reports to avoid any potential misuse of this information that could put a patient in jeopardy. There are indications that some of the drugs perhaps will have valuable pharmacogenetic information available to benefit patients, but still more research is required to confirm the results. Furthermore, the information we looked into was all related to dosing or efficacy. There are other DNA variants listed for these medications related to toxicity, but with poor evidence at the moment. Definitely more research needed!
If you are seeking access to high quality pharmacogenetic testing, contact Merogenomics for what we consider to be one of the best available options on the market. We are big advocates of sequencing your entire genome for a higher investment as a full genome test captures all of the DNA information, including pharmacogenetic related information and everything else. This leaves unlimited future possibilities in terms of how this information can be deciphered from the DNA code, not to mention that upon the receipt of results, the client obtains all the pharmacogenetic interpretation as well as health related information based on the genome.
Of course, just as with pharmacogenetic testing, not all tests are created equal and the same goes for full genome sequencing. So always research your product well and at least make sure they meet the basic minimum quality criteria of DNA testing.
In the meantime, happy genome sequencing!
This article has been produced by Merogenomics Inc. and edited by Jason Chouinard, B.Sc. Reproduction and reuse of any portion of this content requires Merogenomics Inc. permission and source acknowledgment. It is your responsibility to obtain additional permissions from the third party owners that might be cited by Merogenomics Inc. Merogenomics Inc. disclaims any responsibility for any use you make of content owned by third parties without their permission.
Products and Services Promoted by Merogenomics Inc.
Select target group for DNA testing
Healthy screening |
Undiagnosed diseases |
Cancer |
Prenatal |
Or select popular DNA test
 |
 |
 |
 |
Pharmaco-genetic gene panel |
Non-invasive prenatal screening |
Cancer predisposition gene panel |
Full genome |