Cancer mutations and drug resistance
Dr.M.Raszek
Prostate cancer heritability
As we move from October to November, we also move from breast cancer awareness month to Movember which includes raising awareness for prostate cancer. Prostate cancer does not seem to get as much attention as its female counterparts, but that could be because prostate cancer is one of the best managed types of cancers with about a 99% 5-year survival rate!
Consequently, prostate cancer follows a very typical regimen of treatment but the devil can be in details. In this case, the genetic details, as prostate cancers can frequently become resistant to standard therapy approaches, and the genetic information of the cancer can be informative on how such a cancer should be treated.
While perhaps it is still uncommon during a patient’s treatment in Canada to get DNA sequencing, there can be benefits for the right target group. The big argument being: is it worth the investment into a screening method that will be rather expensive to help identify the small minority of patients that would benefit from genetic information?
While the answer to this question with regards to prostate cancer might be more obvious with already high success rates of treatment, the answer really depends on the cancer type. As I pointed out earlier, a large fraction of cancer patients can benefit from DNA sequencing in terms of their medical management, and we are all still acutely aware of how dangerous different forms of cancer can be. Even in cancers that are typically treated quite successfully, there can be situations that are going to be quite dangerous to patient survival, with cancer genetics influencing the severity of the disease outcome to a large degree.
This is the reason why personalized medicine is making such a large impact in current cancer care, and the number of cancer patients who have their DNA analyzed is rising continuously. According to one account, at least 50,000 cancer patients had their genomes sequenced and their information deposited in databases for medical interpretation. No doubt this is only just beginning, as sequencing in cancer care is becoming so prolific, that hundreds of millions of cancer patient genomes are expected to be sequenced by 2030. That is not too far from now, and that is a lot of people! Ask yourself, who in Canada do you know that has already had their genome sequenced for medical purposes, cancer patient or not? As you will soon see, sequencing cancer DNA can provide surprising and powerful information!
But first, let's talk about cancer DNA mutations. In the previous article posted, I discussed the heritability of cancer predisposition. I brought up one study that investigated 33 cancer types among thousands of patients who also had their personal DNA sequenced to see what the rate of inherited cancer promoting mutations is in cancer patients. It turned out that prostate cancer shows a heritable predisposition in approximately 6% of cancer cases, lower than the average of 8% for all cancers. A number of genes can be impacted, including the same genes that are involved in breast and ovarian cancer predisposition (such as BRCA1 and 2), but to a far lesser degree than that seen in women. The prostate cancer predisposition gene that is impacted most frequently, ATM, is also involved in breast and ovarian cancer predispositions. This example drives home the point that many of the same genes can be involved in the development of various types of cancer, and why DNA analysis of cancer can be so important to understand the nature of cancer.
What I did not talk about yet was what these mutations do.
Cancer mutations types
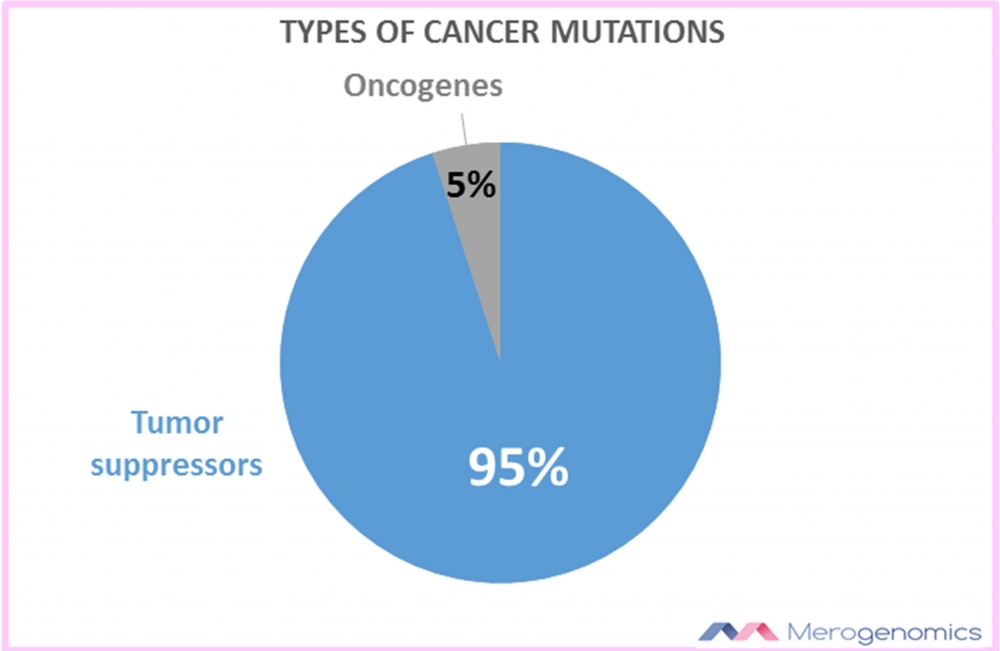
First of all, the majority of cancer mutations happen to be in what we refer to as “tumor suppressors”. What are tumor suppressors? The name sort of gives it away. They are proteins (or as I like to call them, molecular robots), produced in your cells that make sure your cells behave normally. For example, they ensure that cell replication follows a certain pattern. Typically, mutations in the genes that act as the blueprint for the production of these molecular robots end up either completely stopping their production, or producing dysfunctional proteins. This means that the control mechanism to make sure cells divide normally is broken, and cells can suddenly behave abnormally and divide uncontrollably without any response to the surrounding environment. Then you end up with a mass of abnormal cells that can lead to cancer. In other words, when tumor suppressors are broken, certain cellular action that is supposed to be inhibited, is no longer inhibited.
The other type of genes that can be mutated and contribute to cancer development are oncogenes. The oncogenes are on the opposite spectrum of tumor suppressors. With oncogenes, the mutation doesn't break the function of the protein, it enhances it! So what was previously working normally, is now hyperactive and something is done too much. Again, if we go with the idea of cell division, imagine some molecular robot activity was responsible to help cells divide and then became overactive, helping cells divide way more than it is supposed to.
Armed with that information, it is perhaps not surprising then that the majority of mutations are in tumor suppressors and only a minority are in oncogenes. It is, after all, much easier to break something with a random alteration than to enhance it! It reminds me of the story of how corrective eye surgery was discovered: by someone landing on broken glasses and cutting their eyes. Against all odds, that incident improved the vision of that individual!
Judge my mutation by its structure
It may not be surprising to you that in the majority of outcomes of these mutations is reduced activity of these gene products. In other words, these tiny molecular robots are not performing as much activity in cancer cells as would be expected in normal cells. But what might be a surprise to you is how often these mutations that impact the activity of the little robots are large alterations to the genes, such as deletions, a common topic of mine as of late. Once again, the above quoted large study showed that nearly half of all tumor suppressor mutations result in the least active performance of these affected protein robots, and that the majority of these mutations were large truncations of the genes.
To provide you some clue about how common this actually is in the area of medicine, an amazing paper has come out recently based on 143,515 patients that used Invitae panels for a variety of different medical conditions, focusing on the frequency of large structural alterations. This is an enormous data set and an amazing publication on behalf of the company.
What level of frequency of these large alterations are we talking about? In cancer, 8% of all cancer mutations uncovered were these large DNA changes. This included either truncation of information or duplication of genetic information. Another name for these type of alterations is “copy number variation”. For prostate cancer specifically, it is observed slightly less, with about 4% of all prostate cancer patients exhibiting copy number variations.
This was testing all medical conditions, not just cancer, so it is interesting to see this comparison with other conditions. Cardiology showcased approximately a 5% rate of these structural changes. Pediatrics was similar to the cancer rate with 8%. Perhaps the surprising number was in the field of neurology, with an average of 35% of found mutations in such conditions affecting large regions of DNA.
If you want to dig into the genes, BRCA1 showed nearly an 11.5% rate of copy number variations, BRCA2 almost 2%. The other gene that is often found to be a culprit in prostate cancer predisposition, ATM (also one of the top predisposition genes in breast and ovarian cancers), showed a 5.5% rate of large structural DNA changes in this one gene.
This is data collected with gene panels; as I previously pointed out, whole genome sequencing can double the rate of discovery of DNA mutations over gene panels.
Synthetic lethality: beating cancer at its own game
However, DNA sequencing is not merely to provide information on what genes are affected in cancer development, which is important, but rather how can that influence the treatment of the patient. We have been discussing BRCA gene mutations, and patients with these mutated genes have one of the more interesting treatments available to them. BRCA genes produce proteins involved in repairing DNA, specifically repairing DNA that is literally sheared into two pieces. This means that both strands of the DNA double helix must be broken, and such damage is referred to as “double stranded DNA break”.
Imagine a ladder where both of the side rails are broken, so that the ladder is actually divided into two pieces. If only one of the rails was broken, this would be referred to as “nicked” in DNA terminology, or a “single stranded DNA break”. These are insanely common in our cells, but more about that in a moment. The ladder is actually a very good analogy because DNA is typically represented as a ladder with rungs, but imagine if the ladder is twisted along its center axis. Of course such spiral ladders would have no purpose to exist, unless perhaps you are trying to paint two parallel walls in a narrow alley!
How can you fight cancer if one of the primary factors causing the cancer is that the repair mechanism for DNA double stranded breaks is not working? You make sure that there are even more of these breaks in the DNA! Yes, you read that right - we are talking about using inhibitors of single stranded DNA breaks. By increasing the number of single stranded DNA breaks, you increase the likelihood of double stranded DNA breaks. And if you have too many double stranded DNA breaks at the same time, the cell cannot cope with so much damage and dies. Since in this case it is the cancer cells that have this predisposition for double stranded DNA breaks because of the BRCA genes mutations, it is the cancer cells only that are killed in this fashion. This approach of killing cells by causing even more damage is referred to as “synthetic lethality”, and the inhibitors are called PARP inhibitors (for poly ADP ribose polymerase, the enzyme involved in the single stranded DNA break repair). One such PARP inhibitor drug is Olaparib.
Cancer drug resistance
This brings me to the gist of this story which is how cancer cells become resistant.
The current prevailing theory is that drug resistance in cancer is caused by the heterogeneity of cancer cells, meaning that cancer itself is made up of many differently mutated cell types. So that while a specific drug may kill off the bulk of the cancer cells, in this mixture of cancer cells, some cells will already have some resistance to the drug from the start, and eventually will flourish. This is why the current concept for the best cancer treatment option is to use a combination therapy of multiple drugs to ensure that all of the cancer cells are targeted. Such an approach, in theory, should be more effective than any one drug on its own in case the cancer mass were to have some cells that are resistant to one single drug.
There is another possibility that is actually quite fascinating, and that is that cancer cells can mutate themselves in order to acquire resistance. This was demonstrated for the first time against the PARP inhibition not that long ago, in a male breast cancer patient who had inherited cancer predisposition due to truncation of BRCA2 gene, developed breast cancer (very rare in males), and was successfully treated with Olaparib until he developed resistance to the drug. This story is actually a common occurrence in the cancer world. For example, frequently prostate cancers become resistant to androgen deprivation therapy, the main approach of prostate cancer treatment, most often either due to androgen receptor amplification or splicing variants (meaning how the gene product is put together is altered). What is unusual in this case of male breast cancer is that the authors followed up what happened on a molecular level by sequencing the resistant cancer cells’ DNA. What they observed is that the cancer cells had a new mutation in the BRCA2 gene which restored its function to normal! By having a normal function of the BRCA2 gene, the Olaparib treatment with the synthetic lethality approach could no longer work.
The authors were so intrigued by this discovery, that they studied this in another patient with a truncated BRCA2 gene, this time a female, and they observed a similar effect. Once again, the patient was treated with Olaparib to impose synthetic lethality to cancer cells, but they eventually developed resistance. In this case, the mutation that restored the normal function of the gene was even more complex than what was observed with the male patient. In the case of the male patient, it was a single point mutation in the DNA; in the case of the female patient, the mutation involved chromosomal rearrangement to restore the function.
That got me thinking, how did this hypermutagenesis of cancer cells come about? Was it due to random mutations taking place? Cancer cells divide abnormally fast compared to typical healthy tissue, and therefore have a greater chance of random mutations.
Or could this somehow be mechanically activated?
The reason why I had to wonder about that is because of how fast drug resistance develops in cancer patients. In the example of the male patient stated above it only took 10 months. Considering how enormous genome size is, it would seem hard to achieve a hit at the right place through just random mutagenesis. If you recall what I stated earlier, it is much easier to destroy gene function than it is to improve it, so what would be the odds that cancer cells would actually be able to mutate themselves to perfectly restore a specific gene function?
However, the actual amount of DNA lesions that occurs in each cell is actually staggering, approximately 70,000 DNA lesions per cell/day! That is not a typo. That is huge amount of mutagenesis potential! Could such a potential actually be utilized by cancer cells to somehow steer mutagenesis and ensure survival? Sounds like a wild theory, but wilder theories in nature have turned out to be true, and an idea that including single cells, is gaining ground.
Whether true or not, you can see how attractive that notion would be in terms of potential future treatment. If cells somehow can coordinate site specific mutagenesis to elicit a desired response, imagine what that could mean if we could tap into and utilize such an approach. This would mean that we could be mutating cancers back to being a healthy tissue instead of treating them by attempting to kill them (and any other innocent cells that might get in the way). And if that was the case, we wouldn’t have to stop right there, we could be tackling any genetic disease.
This would be in contrast to the current genetic engineering revolution taking place with the CRISPR/Cas9 technologies which requires the invasive introduction of foreign mechanism. Instead we would be using the already present cellular system to execute the process.
It is worth fantasizing about, that’s for sure!
In the meantime, the more cancer patients get sequenced, the more we are going to learn about the potential for treatment. If you need access to cancer DNA sequencing, or want to stay in the loop in this exciting and rapidly advancing field, let us know. Merogenomics will be your strong ally, because if we can beat cancer at its own game, how amazing it would be!
This article has been produced by Merogenomics Inc. and edited by Kerri Bryant. Reproduction and reuse of any portion of this content requires Merogenomics Inc. permission and source acknowledgment. It is your responsibility to obtain additional permissions from the third party owners that might be cited by Merogenomics Inc. Merogenomics Inc. disclaims any responsibility for any use you make of content owned by third parties without their permission.
Products and Services Promoted by Merogenomics Inc.
Select target group for DNA testing
Healthy screening |
Undiagnosed diseases |
Cancer |
Prenatal |
Or select popular DNA test
 |
 |
 |
 |
Pharmaco-genetic gene panel |
Non-invasive prenatal screening |
Cancer predisposition gene panel |
Full genome |