Omicron mutations – battle of the predictions
Dr.M.Raszek
Forest of mutations but what does it mean?
Much has already been said about the Omicron (B.1.1.529) variant thanks to a media onslaught about how infectious this new variant might be, and how many different mutations it has. And it also seemed to come out of nowhere.
But we struggled to find a good summary of the Omicron mutations especially as to why the authorities were so concerned about originally. So, we did bit of our own research.
The first great resource we found was a great database of SARS-CoV-2 mutations from the University of Calgary. It really gives a concise breakdown of what has been previously published on each, already known mutation.
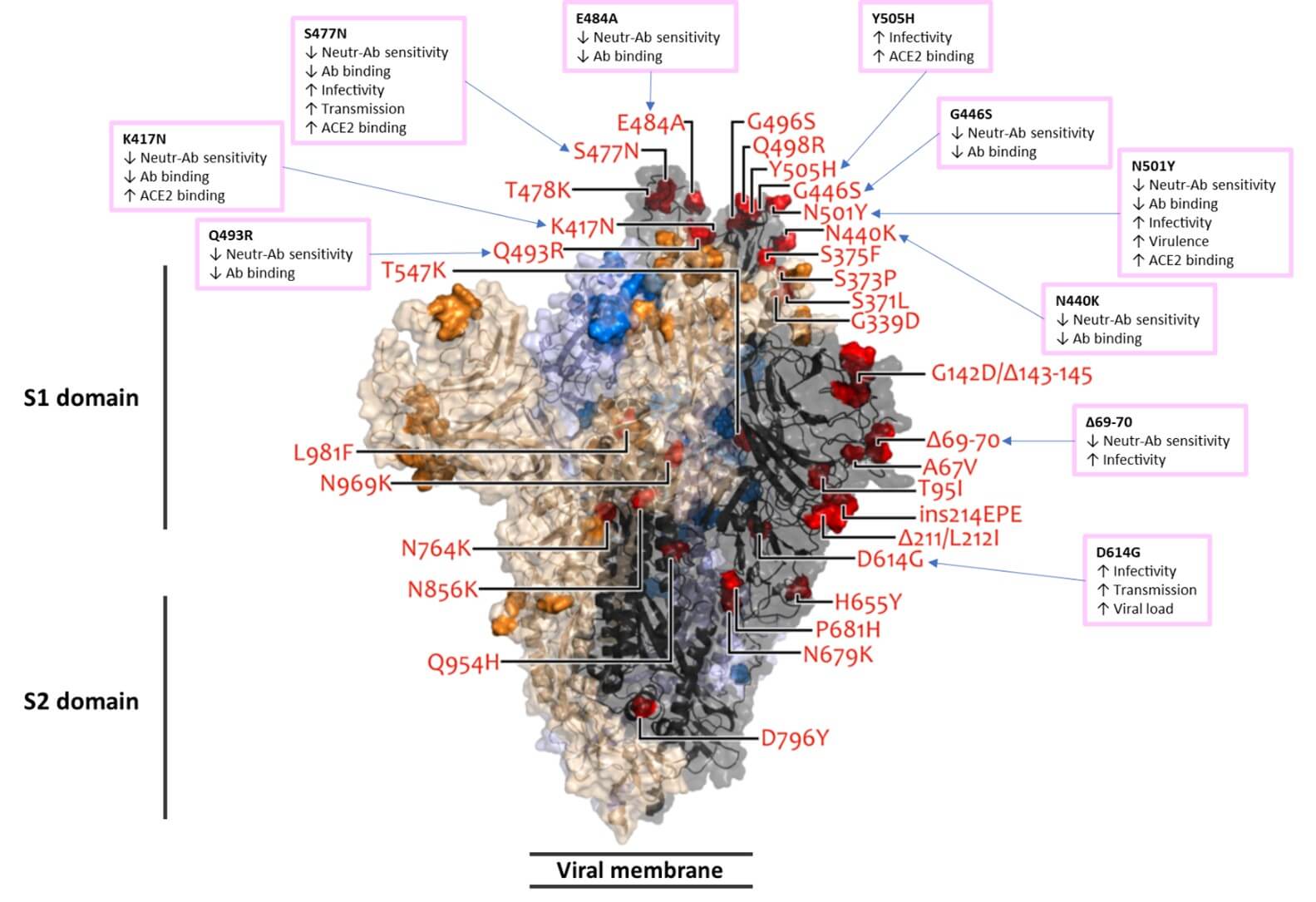
But as we kept on digging into potential consequences, we found some really interesting work from Italy on the possible meaning of these mutations (more on that in a moment). The authors neatly summarized the impact of different Omicron mutations and we wanted to duplicate it here. But we decided to use a different way of demonstrating the location of these mutations by borrowing a great image of Omicron mutations from one of the databases that these authors studied. Below is our own, now favourite, visual summary of Omicron mutations and their impacts.
In the image below, colored red, you can see all of the new mutations of the spike protein alone. Remember, a spike protein is a homotrimer, which means it is made up of three identical units (identical proteins) that make up the final spike protein that is present on the viral surface. These three units are colored, so one unit is dark grey, another is blue and another is beige. The colors help to show you how the three identical units come along together for a final assembly. As a consequence of this trinity, the mutations colored red in the dark grey unit of the homotrimer spike protein, are also present in the remaining two units. They are either dark blue color in the unit represented by blue, or they are colored gold in the beige unit. Labels are provided only for the red mutations in the dark grey unit.
The translucent contours are the expected three-dimensional space of the spike protein, where the shape is determined by the individual amino acids of the protein. Amino acids are the building blocks of proteins. The shape is assigned based on the location of individual atoms that make up these amino acids. This is done using very powerful imaging technologies; thus, we have a profound knowledge of what the spike protein is expected to look like in real life, down to a position for every single atom that makes up the protein. This type of science offers the incredible power to understand and potentially predict outcomes of different mutations. A mutation is a change in the genetic code, which can result in changes in the protein that is built based on the instructions in that genetic code. This is what the mutations in red in the image represent. The contour of the red shape is the contour of the amino acid that occupies that space instead of the original amino acid that was present in the starting Wuhan variant of SARS-CoV-2.
Thus, you get a good idea of what a spike protein looks like as we would expect to see it atop of the SARS-CoV-2 Omicron variant (although do not expect it to display this formidable rainbow of colors).
Underneath that translucent final shape of the protein hides some lines, loops and sheets. They are the backbone of the spike protein and demonstrate a single continuous linear direction of how the string of all the amino acids that make up the spike protein are arranged in space. Remember, when the protein is made based on the genetic code, the protein is in a train of amino acids linked together - one amino acid after another. Eventually these amino acids start interacting with one another (through microscopic interactions based on atomic charges) and that leads to the three-dimensional assembly of the protein. That backbone you are seeing deep inside, traces the three-dimensional arrangement of the continuous line of these amino acids as positioned in space. The amino acids themselves are not shown (except the Omicron specific mutations), but if they were, they would all be attached to that backbone, and their own shape makes up the final translucent shape you are seeing.
If we look closely at just the dark gray colored unit of the spike protein homotrimer, closer to us we see the N-terminal domain (NTD). Further away from us is the receptor binding domain (RBD) which is involved in binding to the ACE2 receptor. Alternatively, if you look at the beige color unit, the N-terminal domain is on the left, while the RBD is on the right, nestling next to NTD and RBD of the dark gray unit. Together, the three RBDs come together at the center top to interact with up to three ACE2 receptors. NTDs are on the periphery, surrounding the center core of the RBDs. NTDs and RBDs together form S1 domain, while the three trunks underneath S1 head form the S2 domain.
Listed in the boxes are some of the most significant impacts of previously observed spike protein mutations in other SARS-CoV-2 variants. These involve impacts on antibodies (Ab) including neutralizing antibodies (Neutr-Ab), the most influential antibodies in the fight against the virus. You can see that quite a few mutations sites have accumulated in the Omicron variant that decrease the binding of antibodies to spike proteins, and thus guess why this inspired worry about this variant. More on that right away. Other impacts of these mutations include potential increases in: infectivity, transmissibility, viral load or ACE2 receptor binding.
But we still have to a great deal to learn about the total impact of many of the other mutations.
And for final bit of clarity, that little triangle you see in some of the names, it denotes the deletion of an amino acid, meaning the amino acid is no longer present in the chain of amino acids that were used to build the protein. Basically, there was a deletion in the genetic code that was used as a template to build the protein. The opposite can also be true, where new amino acids can appear where they previously would not have been present. This is an insertion and is denoted by “ins” in the above image, as the Omicron variant also sports one of those. Once again, there is an insertion of new genetic code in the virus genome, and fascinatingly enough, this insertion is suspected to have originated from a common cold virus. This is covered in Merogenomics first video dedicated to the Omicron variant.
The take home message is that Omicron has lots of mutations that previously have been observed to help the virus escape neutralizing antibodies with the addition of many new mutations that we still do not know what they do. Combine that with the observations on how rapidly Omicron was spreading in the south African region, and it worried health authorities worldwide that we might be dealing with some exceptional variant here. The sheer speed of an Omicron takeover can be seen by the slope below of documented cases for different variants. Omicron just shoots up!
But what can we learn about the new mutations? Is Omicron really more likely to escape antibody neutralization?
Omicron immune escape
This is where the clever work from Italy comes in. But to start this part of the story, first let us define the word epitope. An epitope is any surface area (created by a specific arrangement of atoms) to which an antibody recognizes and binds. An epitope is also any surface area to which receptors of killer cells (like T-cells) recognize and bind. These are the two independent arms of immunity: humoral and cellular immunity. Recognition of the virus is essential in both arms of immunity and that recognition is achieved by physical interaction with the viral proteins. Where those physical interactions take place on virus proteins with our host antibodies or receptors, these regions of contact are called the epitopes.
The Italian team of researchers investigated all known and catalogued epitopes of antibodies/B-cell receptors and T-cell receptors epitopes. This is in a database for all discovered and reported epitopes of any pathogen. In other words, we have a pretty good grasp of how the spike protein is recognized by human biological molecules like antibodies and receptors. This team looked at how many times every single amino acid of the spike protein is recognized by different antibodies/receptors (or another way to put it, how many times each amino acid of the spike protein is part of a different epitope). They plotted this in the form of a graph along with the sequence of spike protein. Basically, the entire spike protein is recognized, and recognized by a multitude of antibodies/receptors that share portions of their epitopes. Thus, every single amino acid in a spike protein participates in multiple, already recognized and studied epitopes. Keep in mind, this is not reflecting what happens in an individual, this reflects what happens in a global population. But there are peaks and valleys, meaning certain areas of the spike protein are definitely more frequently recognized by different antibodies/receptors, than others. That makes perfect sense as this is based on ease of access, (among other things - genetics could also be an influencing factor). Some areas are really easily accessible for molecules to bind to. Some are harder to access. A big chunk of the trunk of the spike protein is not easily accessible for example.
What researchers did then was they counted how often the Omicron mutations affected already known epitopes. And they compared against other known variants of concern. This really clever exercise showed that Omicron mutations affected approximately 30% of both the T-cell and B-cell epitopes. That is a huge impact on the previously characterized attack against the virus. We do not know how these changes will manifest clinically, but many epitopes will be affected. By comparison, Delta had nearly 9% of T-cell epitopes affected and around 11% of the B-cell epitopes affected. Impacting an epitope is not everything though. The virus wants that impact to be translated into lower recognition and hence destruction by the immune system defenses. Prior to Omicron, Gamma variant had the highest distinction of most affected epitopes, with around 12% and 15% of T-cell and B-cell epitopes impacted, respectively. What this means is that higher number is not necessarily better, as Delta had fewer epitopes impacted, but the ones that were affected in Delta had bigger effect for virus benefit than Gamma. But it still points out that Omicron is in some league of its own with the avoidance of epitopes that human biology has targeted to fight the SARS-CoV-2 infection.
Has this been borne out by any evidence yet? Surprisingly yes! Science has been moving at warp speed when it comes to Omicron!
This time the work came from the University of KwaZulu-Natal in South Africa, which might explain why it has been made available so rapidly. The authors showed that in blood of 12 patients who were vaccinated with the Pfizer mRNA vaccine, there was 41-fold reduction in the vaccine’s ability to prevent Omicron virus infection of cells in comparison to an earlier strain that had only the D614G mutation (a strain that emerged early on in the pandemic and took over). This is a dramatic shift and this early experimental evaluation appears to corroborate the potential power of Omicron to bypass vaccines. What was also interesting is that 6 of these patients were also previously infected with the D614G virus besides being vaccinated, and in comparison to vaccinated only, they still exhibited strong protection from the Omicron variant. Weirdly, the authors did not tabulate these independently, so the 41-fold difference drop is for all patients, while for the vaccinated and previously infected patients the drop was 30-fold. This means that for vaccinees only - this number would be even worse but this value was not provided. The authors’ conclusion: “This is estimated to correspond to a vaccine efficacy of 22.5% (95% CI: 8.5%-40.7%) against symptomatic infection, essentially compromising the ability of the vaccine to protect against infection.”
Battle of predictions: Omicron’s impact
Another clue to Omicron expectations comes from a research group in Michigan State University that developed an artificial intelligence analysis of mutational landscape impact. The reason why their work is worth looking into is because they recently predicted the possibility that the SARS-CoV-2 virus will be mutating towards escaping the vaccines, so that is the immune escape we are discussing. And they followed that up with an analysis of the likelihood of an Omicron and vaccine breakthrough. Keep in mind all this work is still so recent, it has not been peer reviewed. But their warning is not trivial. Based on their computational models, the authors suspect that “Omicron may be twice more likely to escape current vaccines than the Delta variant”, as a consequence of which, they estimate it might be twice as infectious as the Delta variant, which would put Omicron in a range of being ten times more contagious than the original Wuhan strain. Their model was based on the amount of binding energy that is attained when antibody/receptor interacts with spike protein epitopes. Think of it as a law of attraction. Stronger binding attraction energy, indicates a stronger interaction. The authors have previously used this model to predict mutations that could provide the SARS-CoV-2 virus an advantage, and such mutations eventually did indeed materialize in future variants. They have been analyzing the possibility of mutations with a potential of immune escape ever since. In this way, the authors predict that Omicron spike protein mutations N440K, T478K, and N501Y should enhance binding of the virus to ACE2 receptor, and therefore enhance infectivity. Additionally, Omicron mutations K417N, E484A, and Y505H are expected to have the largest impact on escaping neutralizing antibodies. You might notice that this is not what one would assume by studying these amino acids changes alone (check above image of the spike protein).
While this might sound dire, it is in direct contrast to another modeling guru group from France. They too have a developed a predictive model, that they call transmissibility index, specifically for SARS-CoV-2. Their claim to fame stems from the fact that using their computational modeling (that also measures binding energies), they successfully predicted that the Delta variant would become the dominant variant in the world. But these authors have an added component in their model, based on their previous proposition that SARS-CoV-2 virus also interacts with specific molecules found on the infected host cell surface (we will just call it a “lipid raft” here where lipids are the primary constituents of the cell membrane, and a raft refers to a congregated collection of such specialized lipids together). We showed their work previously when discussing how antibodies can also contribute to enhancing infection. Their previously calculated transmissibility index for Delta stood at 10.67 versus 2.16 for the starting Wuhan strain, or in other words, 5-fold increase in transmissibility. While transmissibility and infectivity are related but independent events, you can see how this model and that of the group from Michigan are providing similar outcomes for Delta. Where they differ is the predicted outcomes for the Omicron. While the French team did not yet release any official documentation, the team lead, Dr. Jacques Fantini did break the Omicron predictions on social media. With their own model that also incorporates lipid raft interaction with the virus, they expect the Omicron not to be able to outcompete the Delta because Omicron’s calculated transmissibility index measured at 3.90, so almost 3-fold weaker than Delta.
It will be interesting to see which of these titans of variant impact modelling will turn out to be correct.
In the end, time will have to tell us the story of Omicron, but these are some of the types of analyses that are going on behind the scenes if you wonder how these concerns, that the public media loves to amplify to a near psychotic levels, come about in science. The concerns are legitimate, the fear of an unknown tomorrow might not be.
This article has been produced by Merogenomics Inc. and edited by Jason Chouinard, B.Sc. Reproduction and reuse of any portion of this content requires Merogenomics Inc. permission and source acknowledgment. It is your responsibility to obtain additional permissions from the third party owners that might be cited by Merogenomics Inc. Merogenomics Inc. disclaims any responsibility for any use you make of content owned by third parties without their permission.
Products and Services Promoted by Merogenomics Inc.
Select target group for DNA testing
Healthy screening |
Undiagnosed diseases |
Cancer |
Prenatal |
Or select popular DNA test
 |
 |
 |
 |
Pharmaco-genetic gene panel |
Non-invasive prenatal screening |
Cancer predisposition gene panel |
Full genome |