The human genome finally completed!
Dr.M.Raszek
Better late than never
Earlier in the summer, a landmark preprint was published that marks an important milestone in the history of decoding the human genome. For the first time ever, a 100% complete human genome sequence has been decoded and presented!
At Merogenomics, we will relish finally being able to say that a full genome was “fully decoded” as opposed to “almost fully decoded”!
Wait! What? The human genome was not fully decoded already? That’s right, even though the Human Genome Project exhibited the first maps of the human genome in 2000, and announced it as complete in 2003 – a small fraction of the human genome was never resolved due to technological limitations.
Since then, these limitations have been overcome and we have different tools available that can read very long stretches of isolated human DNA. Such technologies are colloquially referred to as long-read sequencing, to differentiate them from the gold-standard DNA decoding technology, which is short-read sequencing. Doesn’t take a genius to figure out that short-read sequencing decodes only short stretches of DNA. In fact, very short, a few hundred bases at most, and usually much less. As a reminder, DNA code is made up of four different bases arranged in a specific manner to act as a storage of instructions. Long read sequencing can obtain a continuous uninterrupted sequence of many thousands of bases long.
Thanks to these new technologies closing that gap of knowledge, the first preprint of an entire human genome sequence ever put together offers some amazing insights!
As this is historic, we have to dive in!
How much human genome was missing?
First, an explanation about this “complete” human genome sequence. When this human genome sequence was finally officially decoded in its entirety, it finished the final gaps that accounted for 8% of the genome that were still missing from previous drafts. This accounted for approximately 200 million base pairs of novel DNA sequences. The authors estimate that this included at least 115 new genes products (remember that genes contain code to direct the production of proteins which then perform specific functions inside our cells once they are made). That is a lot of new genes discovered! Of these new genes, 28 are expected to be of medical impact. For a grand finale of 3,054,815,472 bases in the human genome! The final gene count that codes for the construction of proteins: 19,969.
Who are the authors? They are a consortium of scientists from around the world who banded together to make this historic achievement a final reality. They cleverly named themselves Telomere-to-Telomere (T2T) Consortium. Telomeres mark the ends of chromosomes, which are the large three-dimensional structures into which our massive amount of DNA is woven into. More like spooled into. Super long stretches of DNA are basically super coiled into tighter and tighter packages for compactness. These individual pieces of DNA are called chromosomes. In all, a human genome is divided into 24 pieces called chromosomes, 22 of which are called autosomes (and we all have two sets of it, one from each parent) plus 2 chromosomes called sex chromosomes. We all have two sex chromosomes, again one from each parent, for a total of 46 chromosomes in all of our cells except the reproductive cells which only contains one of the two sets of chromosomes (so 23 chromosomes in total). For the sake of simplicity of this description, we will ignore that there are other combinations as well, for now.
It is the telomeres that are endings to chromosomes, protecting them. Thus, the name of the consortium gives away their goal: obtain a complete sequence of each chromosome from one telomere to another! Basically, all chromosomes still had some gaps that needed to be filled.
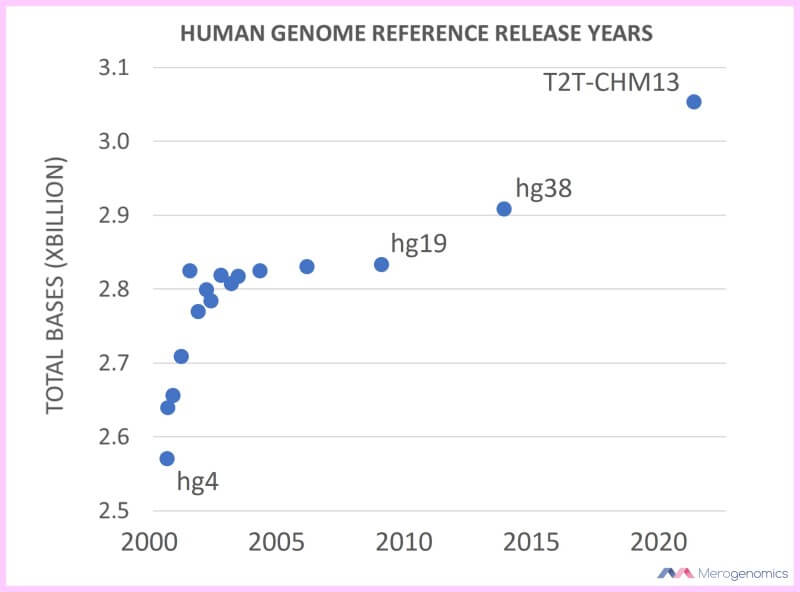
And they finally succeeded! In, 2021, 21 years after first announcement of the human genome sequence and 18 years after first publication of “nearly complete” human genome sequence, while adding the single largest contribution of new content to the human genome in that entire timespan. That was one of our favourite images from that publication, showing how the human genome content has increased over the years in a bid to complete the human genome (not counting mitochondrial genome or the Y sex chromosome).
This is phenomenal for the continuous progress of genomic medicine. Having all completed sequences will allow even greater accuracy in understanding how genetic mutations relate to the production of disease outcomes. A complete sequence automatically removes the guess work if decoded sequences may be an artifact or not (for example, from contaminants or from being misassembled). In essence, the authors produced the most complete and most authoritative human genome reference ever!
Tech that made this magic happen
The currently understood human genome is used as a reference where the DNA code obtained from sequencing someone’s genome is all assigned to very specific locations on the different chromosomes. Use of this reference is absolutely essential for short read sequencing technology and is used all the time in medical genomics. You can dispense with this human genome reference if you use the long read technology, and go for a new attempted assembly from scratch. This is referred to as de novo assembly. This shows you how powerful long read sequencing technologies are in capturing information. Although to build a human genome from scratch without a template to compare to is very hard and requires lots of computational resources.
And if you wonder why they had such a hard time closing these gaps - it is because these sections of DNA were made up of highly repetitive sequences. Imagine a book made up of one repeating sentence. With tiny differences in that sentence here and there (like spelling mistakes). But with so much similarity, how to put a book together of nearly always identical sentences would be very hard if you did not have a template. The reason we finally overcame that is because we finally have technology that can read the entire sequence to tell us how to make that entire book of nearly always identical sentences.
The two long read sequencing technologies employed by the authors, are the only two such leading technologies on the market right now. They are Pacific Biosciences (referred to as PacBio) which still uses polymerase proteins that create a new DNA from a template, and in the process of this DNA duplication, we learn what the code is (because every one of the four bases employed in DNA creation is labelled and specifically recognized when used).
The other technology employed was Oxford Nanopore. This particular technology decodes DNA in a completely different approach. Basically, genetic code is run through a specific miniature pore which electronically senses which of the four bases are running through it. You see, each of the bases has a specific structure to it which means it has a specific electric charge to it. All atoms that make up all matter contribute to an overall electric charge of that matter, however weak or however strong. As a consequence, as a specific base with its own signature electric charge is passing through a pore, it will temporarily affect the electric current that is measured in this place. Each base leaves a specific impact on the current, which can be read as an electronic signature of each base.
Oxford Nanopore brought a completely different way of looking at a sequence, and has changed the game by adding the ability to read the longest continuous sequences of DNA ever, referred to as “ultra-long reads”, even in excess of million bases in length. Just cosmic proportions. But at very high error rate.
Combining Oxford Nanopore ultra long pieces that could act as a template, and PacBio ability to get genetic code in its shorter pieces with very high accuracy did the final magic it seems. Some areas are still a bit iffy, but the authors claim that only 0.3% of the genome now remains in this category.
A new human genome reference guide
But remember how we said there are additional options of how genomic information could be arranged inside a cell?
One such event is when cells that had both maternal and paternal genetic information, then lose the maternal component, and the paternal contribution is duplicated . This is referred to as complete hydatidiform mole (CHM). If that sounds like gibberish, it is because it sure does. But in essence, half of the complexity of the human genome was removed, allowing for easier assembly because the maternal and paternal contributed genomes can vary significantly from one another (because genetic information comes from two separate individuals). Attempting to decode both at the same time could result in mistakes. But this way, it allowed the authors to only deal with one set of parental genomes – the paternal one (don’t worry ladies, you do get the last laugh, below).
These types of events truly do happen in people and can lead to genetic disease development, or the presence of two distinct genetic cell lines making up one individual.
This is why the new full genomic sequence reference is referred by the authors as the T2T-CHM13 reference.
What can we tell you about this all-important new reference?
That the majority of the CHM cell line genome is of European origin and includes regions of Neanderthal origin. Yep, Europeans will never live that one down. Forever stuck with the Neanderthal interbreeding jokes. Perhaps on the bright side, this genetic remnant of 60 thousand years ago or so, shows that we really gave Neanderthals a chance (à la make love not war style).
But jokes aside, the new genome reference did more than just close the gaps. It also identified prior errors in the previous genome reference. Some sequences turned out to be arranged wrong with previous decoding technologies. Over a million bases had to be removed from the previous reference due to false arrangements! 263 genes had to be removed! See ya, gene imposters! Wonder how many more of these we are yet to discover? Yes, clearly, we still have much to be deciphered from the human genome! This complete human genome reference also allows the most accurate way of deciphering complex structural variants, those genetic mutations spanning very large quantities of DNA code. Eventually, as we continue to decode more human genomes with ever increasing levels of accuracy as to how that code is to be accurately arranged, the authors mention the goal of the ultimate creation of a pangenome reference which would then include the full diversity of human genetic variation. We are now a step closer in that direction.
Finally, the new full-length genome is from a single individual. The prior genome reference was a mixture of different individuals with 72.6% of the genome derived from an individual of African-American ancestry (although with some European mixture included), 5.5% of the genome predominantly of East Asian ancestries, while the remaining contributing individuals of predominantly European ancestries. By the way, that reference genome was referred to as GRCh38. Thus, if you already had your genome sequence decoded, you can look at which reference template was used to assemble your genome, and if it is not GRCh38, then it is even an older version, which is prone to an even greater level of inaccurate interpretations. At that point, you might consider reinterpreting your genome sequence. By the way, everyone who has had their own genome decoded, should consider reinterpretation every few years to capture the latest medical interpretations available. Merogenomics specializes in being on the razor’s edge of genomics so book an appointment with us!
The one major limitation of the new T2T-CHM13 reference genome is that it lacks the code of a Y chromosome. Remember how the cell line has a duplicated paternal genome? The sex chromosome that was paternally inherited was an X chromosome. Thus, when the authors used this cell line to decode the full-length human genome, they did it for only the X chromosome. The Y is still missing but they are working on it now. Sorry guys, ladies first.
Future medical impact
This landmark paper did not come out alone. It has been accompanied by a myriad of investigations. One of the more interesting ones looked at the use of this now complete human genome in comparison to previous options, and how that might be impacting interpretations, including T2T-CHM13 full genome medical interpretations. This is the area of interest for Merogenomics as clearly having the most accurate medical interpretation is of highest importance.
To test the impact of this first complete human genome reference, data was compared from a prior 3,202 genomes sequenced with short reads from the famous 1000 Genomes Project (1KGP) that previously looked at the global diversity of human genomes, as well as 17 human genomes from diverse populations that have been sequenced with long reads.
More than 2 million new mutations (variants) were discovered in either the newly accessible regions or inside the corrected regions of the new full genome reference. From a medical point of view, of the previously suspected 4,964 medically-relevant genes in the human genome, 4,924 genes still map to the new T2T-CHM13 reference genome. The new reference genome also improved variant identification and interpretation for 622 medically-relevant genes. This includes reducing incorrect interpretation, and overall, the authors estimated that there was 12-fold reduction in accidental false positives (where you identify a mutation suspected in contributing to a medical condition when it is not really there) in 269 of those medically-relevant genes. That 8% final gap looks like it is going to have pretty good impact on medical genetics.
One of the most important findings concerns the variants listed in the ClinVar database, which catalogues DNA mutations linked to human diseases, and of the 802,674 unique variants in that ClinVar database, 98.4% could still be mapped to the newest T2T-CHM13 full genome reference. And even more importantly, of the 138,927 ClinVar variants that are currently labelled as “pathogenic” or “likely pathogenic” (meaning they are established to contribute towards disease development), which are those that are currently reported to doctors for patient management; 99.5% of these are still mapping to the new reference. This is good because it means there will be limited impact on past medical interpretations that would need new corrections for the interpretation assignments. This makes sense because variants that obtain a label of being “pathogenic” or “likely pathogenic” typically require good quality supporting evidence.
Overall, the authors were very confident in stating, “this genome assembly is poised to replace GRCh38 as the predominant reference for human genetics.”
So, could you buy yourself a complete human genome sequence based on the long reads? Probably yes, although you would have to know where to look and be willing to pay lots of money. We could probably help you get there if that was your ultimate goal in life. This area of genome decoding is still too new to make it economical for regular use, and for a while you will be relegated using short read technology to decode and interpret your genome but having a complete genome for a reference definitely will help from now on in increasing the accuracy of that interpretation.
This article has been produced by Merogenomics Inc. and edited by Jason Chouinard, B.Sc. Reproduction and reuse of any portion of this content requires Merogenomics Inc. permission and source acknowledgment. It is your responsibility to obtain additional permissions from the third party owners that might be cited by Merogenomics Inc. Merogenomics Inc. disclaims any responsibility for any use you make of content owned by third parties without their permission.
Products and Services Promoted by Merogenomics Inc.
Select target group for DNA testing
Healthy screening |
Undiagnosed diseases |
Cancer |
Prenatal |
Or select popular DNA test
 |
 |
 |
 |
Pharmaco-genetic gene panel |
Non-invasive prenatal screening |
Cancer predisposition gene panel |
Full genome |