COVID-19 coronavirus and ethnicity differences
Dr.M.Raszek
Disease predisposition: would you want to know?
Whenever the topic of DNA testing for medical predispositions comes up in a conversation, people’s reaction can be divided into two camps: those who are fascinated by it and would want to learn as much as they could about their health; and those who would rather not know any information, sometimes even if told that such information could help ameliorate the uncovered problem. Many of those who learn that the upfront knowledge could provide a valuable intervention indeed have second thoughts, but nevertheless, there is always a contingent of people who would never want to know, even if this means risking the loss of potential access to an early cure. The argument usually is: the stress of thinking that a disease might or might not materialize would not be worth it.
One of the more compelling questions that has been asked around with regards to the Wuhan coronavirus is whether it infects different ethnic groups to a different extent. This was especially evident before the current global pandemic, when it appeared that Asia was particularly more affected by the virus than the rest of the world.
Conveniently for us, one paper was published recently that attempted to answer that question, and showed that indeed there could be racial differences for coronavirus infection based on what type of genetic mutations different populations might have. So let’s break it down. Hold on to your seats as we are going to descend deep into the scientific rabbit hole! But try to stay with it!
How can we gauge susceptibility to a coronavirus infection from personal DNA?
Very early on it was recognized that the Wuhan coronavirus enters human cells using ACE2 receptors on the surface of our cells.
There are a couple of different ways that DNA mutations can be analyzed to answer the question of such mutations affecting our susceptibility to COVID-19 infection:
- Are there DNA mutations in the ACE2 gene that make the ACE2 receptor less likely to interact with the SARS-CoV-2 coronavirus?
- Are there DNA mutations in and around the ACE2 gene that change the ACE2 receptor quantity on the surface of our cells?
The authors of the linked publication analyzed both. As we pointed out in our last blog post, coronavirus interacts with the ACE2 receptor via a spike protein (also referred to as S-protein), and this interaction has been mapped out in detail. That means we know which amino acids that make up the ACE2 receptor are involved in the interaction with the amino acids of the spike protein found on the surface of the SARS-CoV-2 coronavirus envelope. So the authors had a pretty good clue as to where to look for potential DNA mutations in the gene encoding of the ACE2 receptor. Alas, the answer to the first question was no, we do not have mutations in our population that would reduce the interaction between the receptors on our cells and the coronavirus (with some really rare exceptions). Or as they put it “there was a lack of natural resistant mutations for coronavirus S-protein binding in populations”.
The answer to the second question, however, was a resounding yes.
ACE2 gene expression
First, the authors analyzed a Genotype Tissue Expression (GTEx) database to see what mutations in the ACE2 gene have already been found to affect how the ACE2 gene is used. In other words, which DNA mutations might lead to reduced use of the gene - which in turn would lead to fewer receptors on our cells - and which DNA mutations could result in the opposite or lead to an increased use of the ACE2 gene, with the consequence that more of the ACE2 receptor is produced by our cells, and thus more of the receptor is found on the surface of our cells. The more receptors, then in theory, the more coronavirus binding could occur.
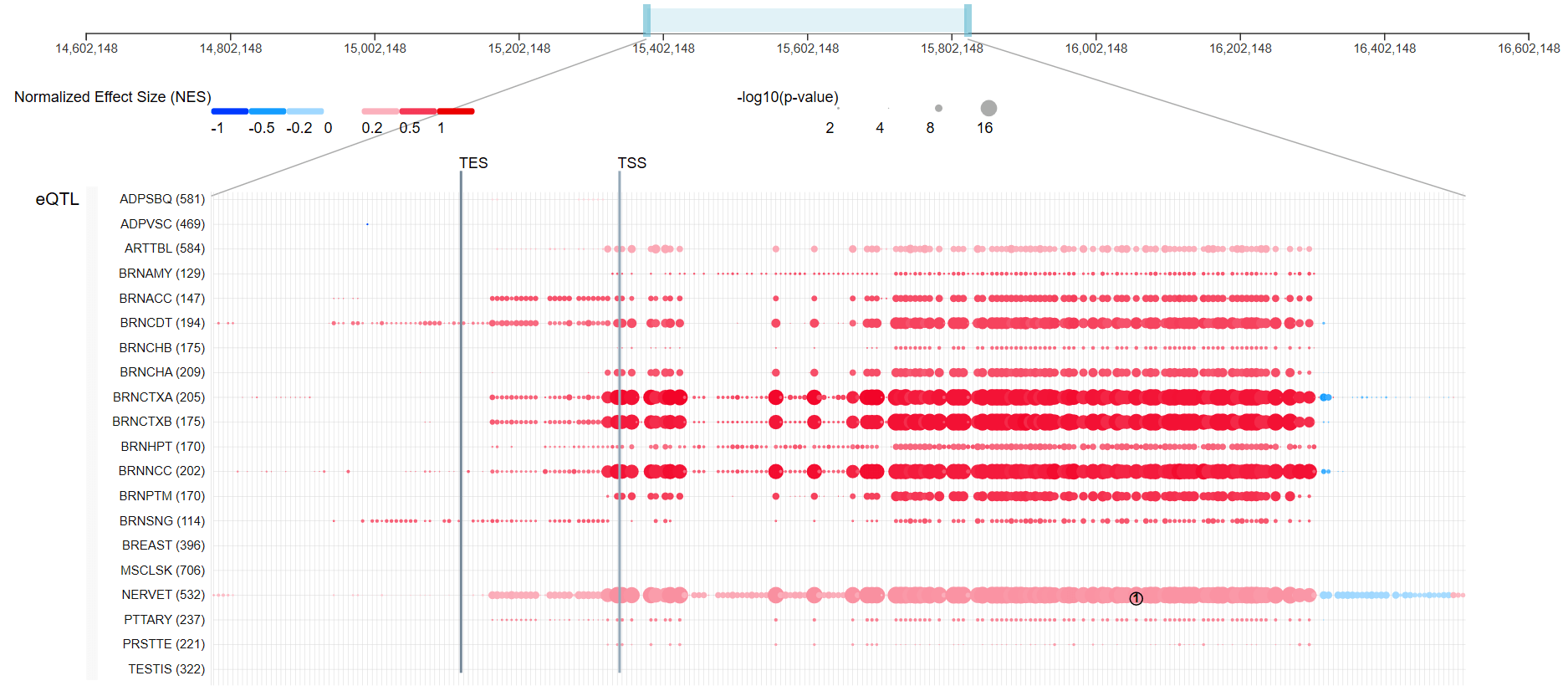
Let’s take a look at this gene ourselves, so you can appreciate what we are talking about.
So what are we looking at? On the very top is a bar showing the gene location within the genome (in this case, the ACE2 gene is present on the X-chromosome, so we are looking at a location along the X-chromosome). The light blue box along that bar is what is displayed below with bunch of red and blue colored dots. Every circle is a data point representing DNA location in and around the ACE2 gene, for which a difference in the gene use (referred to as gene expression) has been previously observed, due to some DNA mutation (or simply, a change in the DNA sequence) in that particular location (jargon for particular DNA location is locus or loci for plural). Just for clarity (or confusion, depending on your mood), these locations are referred to as expression quantitative trait loci, abbreviated as eQTLs.
You can see, there are tons of such putative loci impacting the gene use that have been mapped out. And now imagine, this has been done for thousands of genes! This is an assembly of the work of thousands of individuals all over the world!
Dark blue color represents negative effect (meaning less of the gene is used to produce the receptor) whereas dark red represents positive effect (meaning more of the gene is used to produce the receptor). The size of the circle is in proportion to -log(p-value), or the statistical significance of the observed finding. So in other words, the larger the circle, the less likely that the observed difference seen in the gene expression occurred by mere chance (and therefore the more likely that the effect is real). TSS is the transcription start site while TES is the transcription end site, which is really where, within this entire DNA stretch, is the DNA information that is responsible for eventual production of the ACE2 receptor. For a review of how DNA is used to produce proteins see one of our previous posts. Before the protein can be built, the DNA has to be “transcribed” into a smaller version called RNA (chemically almost identical to DNA) which is then used as a blueprint to produce proteins. The sequence within the DNA and subsequently the RNA, specify which amino acids have to be linked together and in what specific order to produce the protein.
On the left hand side you see some abbreviations for different tissues such as: adipose, arteries, a bunch of different brain tissues, breasts and mammary tissue, skeletal muscle, nerve tissue, pituitary, prostate and testis.
As you can see, there are tons of DNA mutations that have been mapped out that lead to the increased production of the ACE2 receptor - the receptor used by the coronavirus. You can also note that the majority of these mutations lie outside the ACE2 gene itself, in the region of the DNA that is typically used for the regulation of gene activity. There are some DNA mutations within the gene, but all of these are in fact inside introns, and none within exons. It is the exon components of the gene that end up being used together to produce the RNA transcript that is subsequently used as the blueprint for the production of the receptor.
It is these DNA mutations we can examine and ask: are they different in different ethnicities?
Ethnic differences in ACE2 gene use
Since there are so many mutations, the authors compiled a list of DNA mutations (also referred to as variants in more proper scientific jargon) with the largest statistically significant effect size in the ACE2 gene expression, and then they combed through additional, different databases to see how frequently these variants were responsible for this increased (or decreased) production of the ACE2 receptor being observed in different ethnicities.
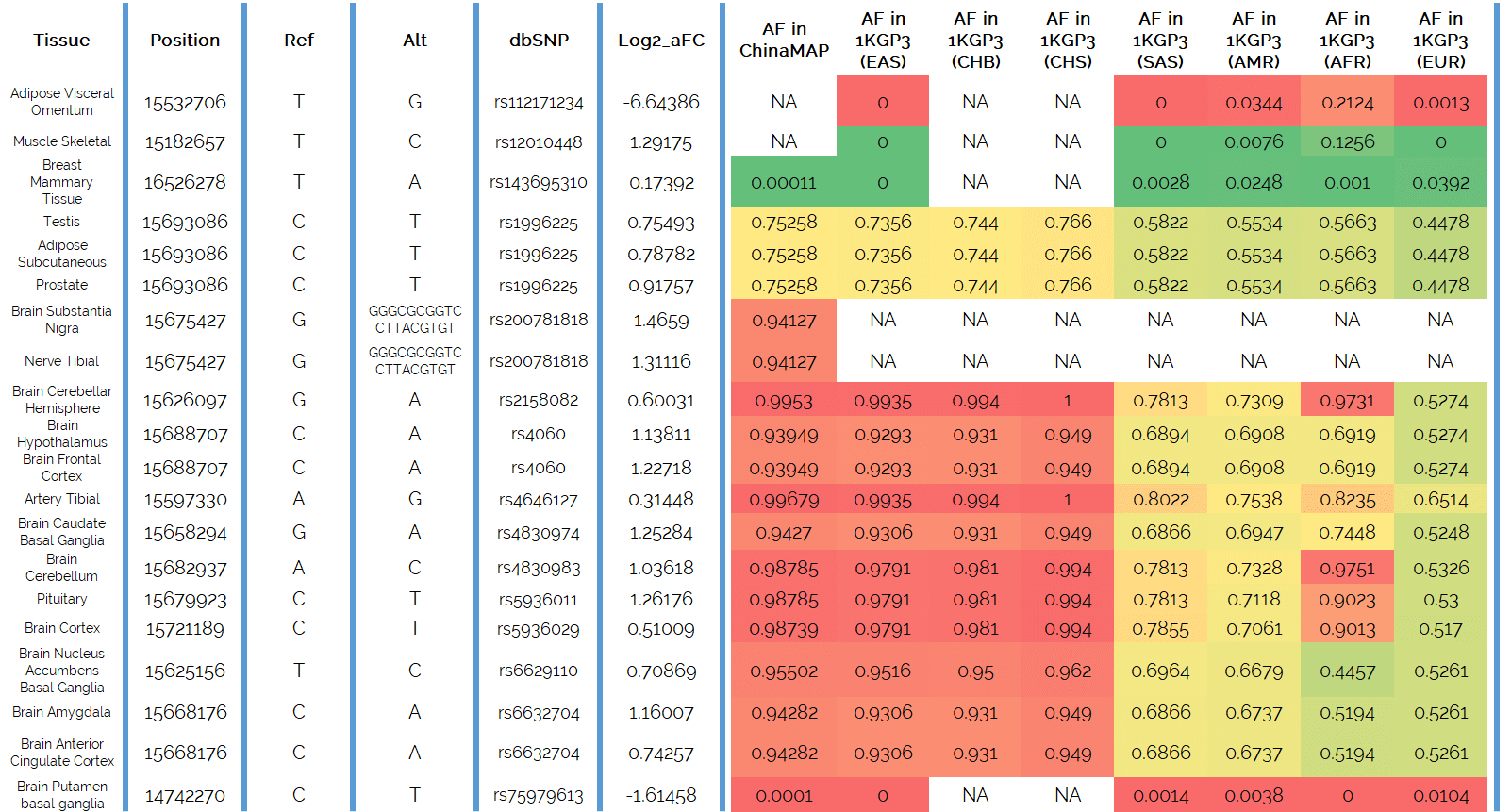
Here is what they found (do not panic, an explanation follows):
The first column lists the tissues from which the data on differences of ACE2 gene expression was derived. Second is the actual nucleotide position on the X-chromosome that is the location where the different DNA variants are observed that are responsible for the different use of the ACE2 gene. We wanted to include that for those who have their genome sequence available to them and were simply curious to see what their own DNA sequence is. The third column is the nucleotide that is observed in the human reference genome and we use it as a reference to compare against. This does not mean that it is the predominant version found in a population, or that it is ideal nucleotide for human health. It is simply what was found when the very first human genome was decoded and is now a reference to compare against.
The fourth column is the alternative variant that can be observed in different people in that same location. The fifth column is the code name for that specific alternative variant for that particular location in the genome. Every nucleotide location in the human genome that has an alternative option found in the human population, such alternative variant has a specific identification code. If you sequence your genome to screen for medical predispositions, the report your doctor will receive will always include the affected gene, the DNA variant found and its identification number such as these, on top of what condition was identified.
Next we get to the good stuff. The sixth column, labelled “log2_aFC”, denotes what is called the log allelic fold change. In essence, this column shows you how much the DNA change affects the use of the gene. It shows how different the ACE2 gene expression (in other words: its use) is if we take the expression level for the gene with alternative nucleotide version in the gene over the expression level for the gene if the nucleotide is same as that of the reference genome. Plus a reminder, you inherited this same gene from each of your parents, so you have two copies in your genome: one from mom, one from dad. What we are comparing here is if both of the inherited genes have the same nucleotide present. This is called homozygous state. So we are comparing here individuals who were homozygous for the alternative nucleotide in a specific location of the ACE2 gene to those homozygous for the nucleotide that is the same as in the reference genome.
Whatever that ratio difference is, it is expressed as a binary logarithm. This is done so that we can see what the fold difference is between the alternate DNA variant and the reference genome variant, which is simpler to use. Thus, if you see a positive number, that is how many fold different the group with increased ACE2 gene use is over the other. If you see a negative number, that is how many fold that same group is lower in ACE2 gene use over the other. If you wanted to get the actual ratio, you would take 2 to the power of whatever value is listed. But that would mean that if the ACE2 gene use is lower in comparison to the reference, an under-expressed ACE2 gene will have values between 0 to 1 (because we are now showing that it is only a fraction of the reference). But an over-expressed ACE2 gene can have values from 1 to infinity. Hence the use of binary logarithm to express the fold-difference.
This means that for the higher the positive value listed, the alternative nucleotide has a larger expression of the ACE2 gene than the reference one, and the number is the fold difference between the two. Conversely, the higher the negative value listed then the reference genome nucleotide has a larger expression of ACE2 gene than the alternative nucleotide.
Where does that lead us? Into looking at which ethnicities have the alternate DNA variants that increase the ACE2 gene expression.
All the remaining columns indicate how frequently the alternate genome nucleotide is found in different ethnic populations: AFR = African; AMR = Ad Mixed American (South and Central Americas); CHB = Han Chinese in Beijing; CHS = Han Chinese South; EAS = East Asian; EUR = European; SAS = South Asian.
For further clarification, the 1KGP is the 1000 Genomes Project database that lists these population frequencies of different mutations across the human genome. ChinaMAP is a China Metabolic Analytics Project which compiles even more granular data within specific populations of the world, and is shown for a comparison as it includes data on one of the unique DNA alterations that results in a small sequence insertion in the genome. These type of DNA alterations can be on a much grander scale, involving thousands of nucleotides. These are referred to as DNA structural variants. By comparison, this is peanuts. When such insertions or deletions are under 50 nucleotides, they are nicknamed indels, and they are very common in our genomes.
The color scheme is to show that the lower the DNA variant frequency (termed allele frequency, hence the AF abbreviation) of a variant that increases the expression of the ACE2 gene (and therefore presumably higher levels of ACE2 receptor on the surface of cells), the greener the color (because that is what we want). The higher the frequency of such variants that increase the expression of the ACE2 gene, the redder the color (not a favourable condition).
Right away you can see that the East Asian population EAS which comprises CHB and CHS Chinese populations, is worst off in terms of the frequency of mutations that increase the ACE2 gene expression. Caucasians appear to be best off with lowest frequency of such mutations.
But to really know where you stand, you would have to look at your genome sequence to see if you have the predisposing nucleotides or not.
Of course, you could be confused as to the group definitions, so here is the description of the actual populations studied in the 1000 Genomes Project:
| Population | Code | ||
| African ancestry | AFR | ||
| Esan in Nigeria | ESN | ||
| Luhya in Webuye, Kenya | LWK | ||
| Mende in Sierra Leone | MSL | ||
| Yoruba in Ibadan, Nigeria | YRI | ||
| African Caribbean in Barbados | ACB | ||
| People with African Ancestry in Southwest USA | ASW | ||
| Americas | AMR | ||
| Colombians in Medellin, Colombia | CLM | ||
| People with Mexican Ancestry in Los Angeles, CA, USA | MXL | ||
| Peruvians in Lima, Peru | PEL | ||
| Puerto Ricans in Puerto Rico | PUR | ||
| East Asian ancestry | EAS | ||
| Chinese Dai in Xishuangbanna, China | CDX | ||
| Han Chinese in Beijing, China | CHB | ||
| Southern Han Chinese Southern Han | CHS | ||
| Japanese in Tokyo, Japan | JPT | ||
| Kinh in Ho Chi Minh City, Vietnam | KHV | ||
| European ancestry | EUR | ||
| Utah residents (CEPH) with Northern and Western European ancestry | CEU | ||
| British in England and Scotland | GBR | ||
| Finnish in Finland | FIN | ||
| Iberian Populations in Spain | IBS | ||
| Toscani in Italia | TSI | ||
| South Asian ancestry | SAS | ||
| Bengali in Bangladesh | BEB | ||
| Gujarati Indians in Houston, TX, USA | GIH | ||
| Indian Telugu in the UK | ITU | ||
| Punjabi in Lahore, Pakistan | PJL | ||
| Sri Lankan Tamil in the UK | STU |
The face value of the data thus appears to show that certain ethnicities are worse off just because they have extra amount of some receptor. However, is this simply an unfortunate biological byproduct? Obviously this receptor plays an important biological function and in fact, under normal circumstances, it might even provide some biological advantages. ACE2 is a receptor for angiotensin which is a hormone that narrows your blood vessels. This leads to increased blood pressure forcing your heart to work harder, allowing you to kick some ass. Another potential benefit ironically, is that ACE2 might protect lungs from inflammatory injury. The irony is that the deadliest danger of contracting COVID-19 is dying from an acute respiratory distress syndrome caused by an extreme inflammation of the lung tissue. So here we have an evil twist that the coronavirus injures us by using the very receptor that otherwise could be protecting us from the very damage that the coronavirus causes.
Let's come clean with the lungs
But if you have been a sharp observer, you might have noticed that none of the tissues examined were actually lung tissue, so how relevant is this really? Perhaps it does not really matter because the pattern that we observe here in different tissues does not hold for lungs where we expect the coronavirus to do its damage. That could very well be true.
But as we pointed out in our previous post on the Wuhan coronavirus structure, ACE2 receptors used by SARS-CoV-2 are also used by SARS-CoV virus, and therefore ACE2 tissue distribution has been previously studied. Lungs definitely express it, in fact in abundance, and we know exactly which cells in the lungs express it. We just don’t know its expression difference based on ethnicity.
Well, some scientists in China recently looked into just that, and published a preprint (research results posted prior to peer review) on expression of ACE2 gene in lungs. These authors looked at ACE2 gene expression by looking at the ACE2 RNA amounts (one of the ways of measuring gene use, another would be looking at protein amount) in various lung cells of 8 individuals: 5 African American, 2 white and 1 Asian. The only conclusion provided on the ethnicity analysis was that “Asian donor (male) has a much higher ACE2-expressing cell ratio than white and African American donors (2.50% vs. 0.47% of all cells)”. Or in other words, way more, enough for authors to label it an “extremely large number”.
Thus it appears that ACE2 gene expression in lung tissues also might exhibit ethnic differences, just as the other tissues we investigated here. Whether any of the listed variants will turn out to be significant for enhanced infection by the SARS-CoV-2 coronavirus will have to await proper scientific investigation.
For now, at least we know what the DNA variants that might influence the number of receptors for the coronavirus are, and so they can be individually investigated.
Should you alter your behaviour if you find you have DNA variants that might predispose you to increased ACE2 receptor production?
Absolutely! Because the behaviour changes, which is the ultimate purpose behind analyzing your own genome, in this instance are simple and smart to do no matter what.
What can you do:
- Increase your hygiene standards dramatically!
- Individual isolation as much as possible
- Increase the radius of public cleanliness nearest to you (especially surface areas touched by hands)
Adhere to these steps as much as life circumstances allow you, until the pandemic ends.
Help others as much as possible while maintaining minimal contact. Top priority for those who will need help will be our doctors and nurses. In pandemics they are our most precious resource and we need to protect their well being. Next are those people who will be forced to quarantine themselves, which can be a very difficult experience.
Genetics or otherwise, what needs to be achieved is to minimize the spread of the virus so that the virus can run out its life cycle and the pandemic can die out. For that to be achieved, we need to either stop it through molecular intervention (not available yet) or preventing host access (ourselves).
If genetics is what you need to persuade yourself, so be it. But you also do not need it. You can take proactive steps without it, and we recommend you do, especially since we do not know the practical validity of this type of genetic information, yet.
Still, we go back to the original question, would you want to know if you were more vulnerable based on validated genetic information? What group do you belong to: those who want to know up front about their potential predispositions or those who rather not know?
This article has been produced by Merogenomics Inc. and edited by Jason Chouinard, B.Sc. Reproduction and reuse of any portion of this content requires Merogenomics Inc. permission and source acknowledgment. It is your responsibility to obtain additional permissions from the third party owners that might be cited by Merogenomics Inc. Merogenomics Inc. disclaims any responsibility for any use you make of content owned by third parties without their permission.
Products and Services Promoted by Merogenomics Inc.
Select target group for DNA testing
Healthy screening |
Undiagnosed diseases |
Cancer |
Prenatal |
Or select popular DNA test
 |
 |
 |
 |
Pharmaco-genetic gene panel |
Non-invasive prenatal screening |
Cancer predisposition gene panel |
Full genome |